Instructions
Objective
Write a MIPS assignment program to search DNA sequence in and C.
Requirements and Specifications
DNA Search: This project explores pattern matching techniques to find a pattern in a DNA sequence containing letters in the DNA alphabet {A, C, G, T}. For example, suppose we have a DNA sequence as follows:
ATGACGATCTACGTATGGCAGCCACGCTTTTGATGTTAAGTCACACAGCCAAGTCAACAAGGGCGACTTCATGATCTTTCCGCTCCGTTGGTGTAGGCCCGTGTTCAAATTCAATGGCTGATTGGAATTACCTTTGAAATACTCCAACCGACCGCCACGGCCAGGGTCCCGCTCGCTCTCTGTGGCCCTCCCACAAAACTCCGGTGAAAGTTGATTTGGACACGGACCCAAAGCAGCGTAGATTATTCGAGCGTATTCGGTAGTCATTGAGGCCCCAA
The pattern “GCTTTT” can be found at index 27 (where the first letter of the sequence is at index 0). Note that overlapping matches are treated individually. For example, if the sequence is ‘AAAAAA’ and the pattern is ‘AAA’, there are 4 occurrences of the pattern: at indices 0, 1, 2, and 3. You will write a C program and a MIPS program to find the indices at which a given pattern occurs in a given DNA sequence.
Strategy: Unlike many “function only” programming tasks where a solution can be quickly envisioned and implemented, this task requires a different strategy.
- Before writing any code, reflect on the task requirements and constraints. Mentally explore different approaches and algorithms, considering their potential performance and costs.The metrics of merit are static code length, dynamic execution time, and storage requirements. There are often trade offs between these parameters.
- Once a promising approach is chosen, a high level language (HLL) implementation (e.g.,in C) can deepen its understanding. The HLL implementation is more flexible and convenient for exploring the solution space and should be written before constructing the assembly version where design changes are more costly and difficult to make. For P1-1, you will write a C implementation of the program.
- Once a working C version is created, it’s time to “be the compiler” and see how it translates to MIPS assembly. This is an opportunity to see how HLL constructs are supported on a machine platform (at the ISA level). This level requires the greatest programming effort; but it also uncovers many new opportunities to increase performance and efficiency. You will write the assembly version for P1-2.
Screenshots of output
Source Code
C language
/*
ECE 2035 Project P1-1
Please fill in the following
Student name:
Current date:
This is the only file that should be modified for the C implementation
of Project 1.
This program initializes a DNA sequence of 10,240 random characters and
a pattern of 3 to 7 random characters, all characters are from the DNA
alphabet {A, T, G, C}.
For Project P1-1, you will implement the function Match, called by main.
The function is defined at the end of this file.
*/
#include
#include
#include
// DO NOT include any additional libraries.
#define DEBUG 0 // RESET THIS TO 0 BEFORE SUBMITTING YOUR CODE
#define SEQLEN 10240
#define MAXPATLEN 10
#define NUMCHAR 4
char Alphabet[] = "ACTG";
int MatchIndices[SEQLEN];
int main(int argc, char *argv[]) {
char Seq[SEQLEN], Pat[MAXPATLEN];
int I, PatLen;
void Print_Seq(char *Seq, int SeqLen);
void Print_Pat(char *Pat, int PatLen);
void Match(char *Pat, int PatLen, char *Seq, int SeqLen);
if (DEBUG){
printf("Sample debugging print statement.\n");
}
srand((unsigned int) time(NULL)); // seed random number generator
PatLen = (rand() % MAXPATLEN) + 1; // compute pattern length
for (I = 0; I < SEQLEN; I++)
Seq[I] = Alphabet[rand() % NUMCHAR]; // create sequence
for (I = 0; I < PatLen; I++)
Pat[I] = Alphabet[rand() % NUMCHAR]; // create pattern
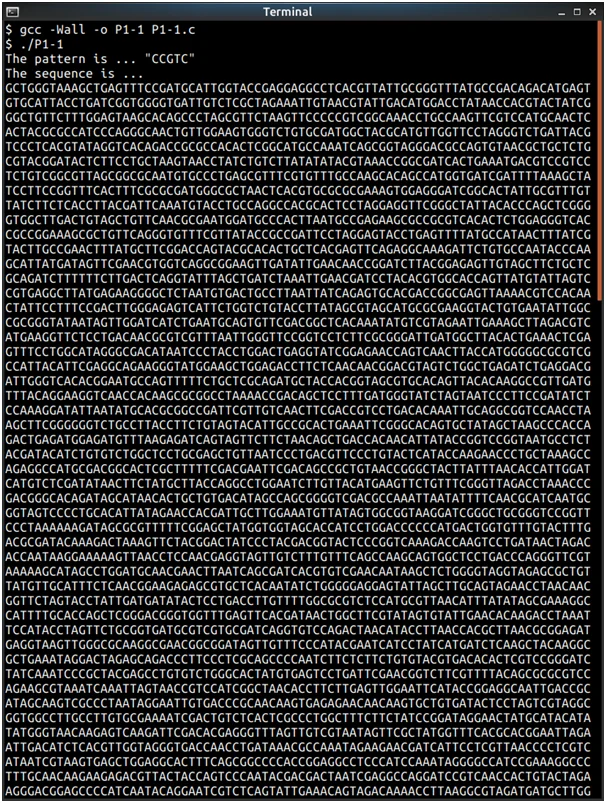
Print_Pat(Pat, PatLen); // print pattern
Print_Seq(Seq, SEQLEN); // print sequence
Match(Pat, PatLen, Seq, SEQLEN); // match pattern in sequence
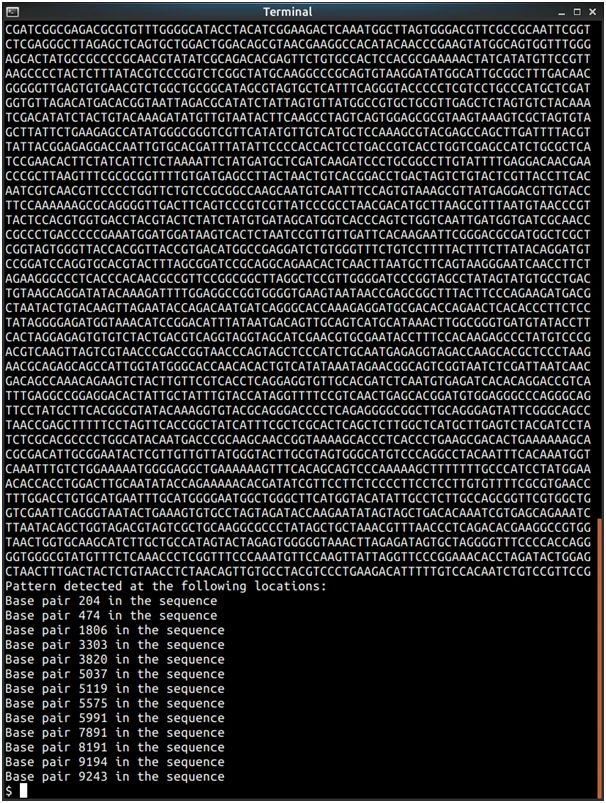
printf("Pattern detected at the following locations:\n");
I = 0;
while (MatchIndices[I] != -1)
printf("Base pair %d in the sequence\n", MatchIndices[I++]);
return 0;
}
/* Print Sequence
This routine prints the sequence. */
void Print_Seq(char *Seq, int SeqLen) {
int I;
printf("The sequence is ...\n");
for (I = 0; I < SeqLen; I++) {
putchar(Seq[I]);
if (I % 80 == 79)
printf("\n");
}
}
/* Print Pattern
This routine prints the match pattern. */
void Print_Pat(char *Pat, int PatLen) {
int I;
printf("The pattern is ... \"");
for (I = 0; I < PatLen; I++)
putchar(Pat[I]);
printf("\"\n");
}
/* Match
This routine find indices of occurrences of a variable length DNA
pattern in a DNA sequence and stores them in the global array MatchIndices.
It stores a -1 at MatchIndices[n] (where n = number of occurrences of the pattern) to indicate the end of the sequence of indices. */
void Match(char Pat[], int PatLen, char Seq[], int SeqLen) {
// insert your code here
MatchIndices[0] = -1; // replace this
return;
}
Assembly language
# DNA Search
#
# This program computes the indices of all (possibly overlapping) occurrences
# of a pattern string in an input DNA sequence, given in the array starting
# at the label Input. It stores the indices in the output array starting at
# label Indices.
#
# Please fill in the following
# Student name:
# Date:
.data
Input: .alloc 600
Indices: .alloc 4800
.text
DNAsearch: addi $1, $0, Input # point to input base
swi 548 # create DNA search
# your code goes here
addi $1, $0, 5 # highlight position
swi 549
addi $1, $0, 86 # highlight position
swi 549
addi $1, $0, 167 # highlight position
swi 549
addi $2, $0, 1 # store the output
swi 580 #
jr $31 # return to caller
Similar Samples
Discover top-notch programming solutions with our expertly crafted samples at programminghomeworkhelp.com. Whether you're tackling complex algorithms or debugging code, our examples showcase the quality and depth of our assistance. Get inspired and see how we can help elevate your programming projects.
C
C
C
C
C
C
C
C
C
C
C
C
C
C
C
C
C
C
C
C