Instructions
Objective
Write a python assignment to implement stochastic gradient descent.
Requirements and Specifications
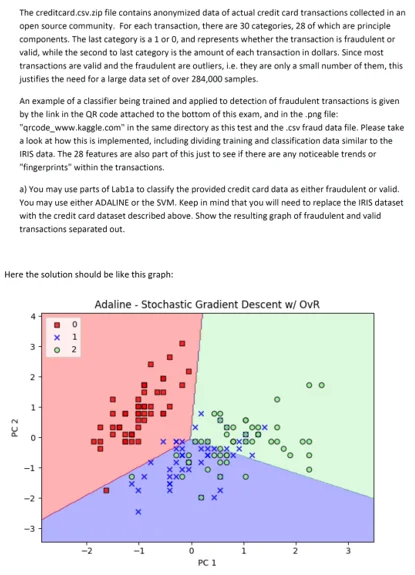
Source Code
!pip install --upgrade --no-cache-dir gdown
## Download Dataset from drive link
!gdown --id 1H7ONGAS2hZgOBIq8csIdjjpNGVs4aWPL
import pandas as pd
import numpy as np
from sklearn import preprocessing
from sklearn.metrics import confusion_matrix
from sklearn import svm
import seaborn
import matplotlib.pyplot as plt
from sklearn.metrics import plot_confusion_matrix
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import accuracy_score
from sklearn.ensemble import RandomForestClassifier
from sklearn.decomposition import PCA
## Load Dataset
df = pd.read_csv('creditcard.csv')
df = df.astype({col: 'float32' for col in df.select_dtypes('float64').columns})
df = df.astype({col: 'int32' for col in df.select_dtypes('int64').columns})
df.head()
## Normalize Dataset
scaler = MinMaxScaler(feature_range=(0, 1))
normed = scaler.fit_transform(df)
df_normed = pd.DataFrame(data=normed, columns=df.columns)
df_normed.head()
## Describe
df.describe()
## Check correlation between variables
plt.figure()
seaborn.heatmap(df.corr(), cmap="YlGnBu") # Displaying the Heatmap
#seaborn.set(font_scale=2,style='white')
plt.title('Heatmap correlation')
plt.show()
# Balance
df_1 = df[df['Class'] == 1]
df_0 = df[df['Class'] == 0].iloc[:len(df_1),:]
df = df_0.append(df_1, ignore_index = True)
df = df.sample(frac=1)
scaler = MinMaxScaler(feature_range=(0, 1))
normed = scaler.fit_transform(df)
df_normed = pd.DataFrame(data=normed, columns=df.columns)
df_normed.head()
## Split into train and test
train = df_normed.sample(frac=0.7)
val = df_normed.loc[~df_normed.index.isin(train.index)]
train.reset_index(drop=True, inplace=True)
val.reset_index(drop=True, inplace=True)
## Split data into X and y
y_train = train['Class']
X_train = train.drop(columns = ['Time', 'Amount', 'Class'])
y_val = val['Class']
X_val = val.drop(columns = ['Time', 'Amount', 'Class'])
## PCA
pca = PCA(n_components = 2)
pca.fit(X_train)
X_train = pca.transform(X_train)
pca = PCA(n_components = 2)
pca.fit(X_val)
X_val = pca.transform(X_val)
## Create Model
model = svm.SVC(kernel = 'linear',C=1.0)
model.fit(X_train, y_train)
## Measure Accuracy
y_pred = model.predict(X_val)
model_acc = accuracy_score(y_val, y_pred)
print(f"The accuracy of the model is: {model_acc}")
## Confusion matrix
fig, axes = plt.subplots(nrows = 1, ncols = 2, figsize=(8,8))
plot_confusion_matrix(model, X_train, y_train, ax = axes[0])
plot_confusion_matrix(model, X_val, y_val, ax = axes[1])
plt.show()
## Display clustering
x_min, x_max = X_train[:, 0].min() - 1, X_train[:, 0].max() + 1
y_min, y_max = X_train[:, 1].min() - 1, X_train[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
plt.figure(figsize=(8,8))
ypred = model.predict(np.c_[xx.ravel(), yy.ravel()])
ypred = ypred.reshape(xx.shape)
plt.contourf(xx, yy, ypred, cmap = plt.cm.coolwarm, alpha = 0.8)
idx = np.where(y_train == 1)[0]
plt.scatter(X_train[idx,0], X_train[idx,1], c = 'red',cmap = plt.cm.coolwarm, marker='o',edgecolors='black', label = '1')
idx = np.where(y_train == 0)[0]
plt.scatter(X_train[idx,0], X_train[idx,1], c = 'blue',cmap = plt.cm.coolwarm, marker='s',edgecolors='black', label = '0')
plt.legend()
plt.legend()
# Part 2) Now cluster for each pair of consecutive features
fig, ax = plt.subplots(nrows = 2, ncols = 14, figsize=(30,10))
j = 0
k = 0
for i in range(27):
X_train = train.drop(columns = ['Time', 'Amount', 'Class']).iloc[:,i:i+2]
X_val = val.drop(columns = ['Time', 'Amount', 'Class']).iloc[:,i:i+2]
model2 = KNeighborsClassifier(n_neighbors=2)
model2.fit(X_train,y_train)
y_min, y_max = X_val.values[:, 1].min() - 1, X_val.values[:, 1].max() + 1
x_min, x_max = X_val.values[:, 0].min() - 1, X_val.values[:, 0].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
ypred = model2.predict(np.c_[xx.ravel(), yy.ravel()])
ypred = ypred.reshape(xx.shape)
ax[j,k].contourf(xx, yy, ypred, cmap = plt.cm.coolwarm, alpha = 0.8)
idx = np.where(y_val == 1)[0]
ax[j,k].scatter(X_val.values[idx,0], X_val.values[idx,1], c = 'red',cmap = plt.cm.coolwarm, marker='o',edgecolors='black', label = '1')
idx = np.where(y_val == 0)[0]
ax[j,k].scatter(X_val.values[idx,0], X_val.values[idx,1], c = 'blue',cmap = plt.cm.coolwarm, marker='s',edgecolors='black', label = '0')
ax[j,k].axis('off')
ax[j,k].legend()
ax[j,k].set_title(f'V{i+1} vs. V{i+2}')
k+=1
if k%14 == 0:
j += 1
k = 0
plt.show()
Now, we see that since we only have two classes, the best number of clusters/neighbors to select is 2. We see how each pair of variables is clustered in the multi-plot figure shown above.
# Random Forest
fig, ax = plt.subplots(nrows = 2, ncols = 14, figsize=(30,10))
j = 0
k = 0
for i in range(27):
X_train = train.drop(columns = ['Time', 'Amount', 'Class']).iloc[:,i:i+2]
X_val = val.drop(columns = ['Time', 'Amount', 'Class']).iloc[:,i:i+2]
model3 = RandomForestClassifier(max_depth=2, random_state=0)
model3.fit(X_train,y_train)
y_min, y_max = X_val.values[:, 1].min() - 1, X_val.values[:, 1].max() + 1
x_min, x_max = X_val.values[:, 0].min() - 1, X_val.values[:, 0].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
ypred = model3.predict(np.c_[xx.ravel(), yy.ravel()])
ypred = ypred.reshape(xx.shape)
ax[j,k].contourf(xx, yy, ypred, cmap = plt.cm.coolwarm, alpha = 0.8)
idx = np.where(y_val == 1)[0]
ax[j,k].scatter(X_val.values[idx,0], X_val.values[idx,1], c = 'red',cmap = plt.cm.coolwarm, marker='o',edgecolors='black', label = '1')
idx = np.where(y_val == 0)[0]
ax[j,k].scatter(X_val.values[idx,0], X_val.values[idx,1], c = 'blue',cmap = plt.cm.coolwarm, marker='s',edgecolors='black', label = '0')
ax[j,k].axis('off')
ax[j,k].legend()
ax[j,k].set_title(f'V{i+1} vs. V{i+2}')
k+=1
if k%14 == 0:
j += 1
k = 0
plt.show()
Similar Samples
Discover a wide range of programming homework samples at Programming Homework Help. Our examples showcase expert solutions to complex coding challenges, helping you understand various programming concepts and improve your coding skills. Get inspired and learn how to tackle your assignments effectively!
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
Python
.webp)